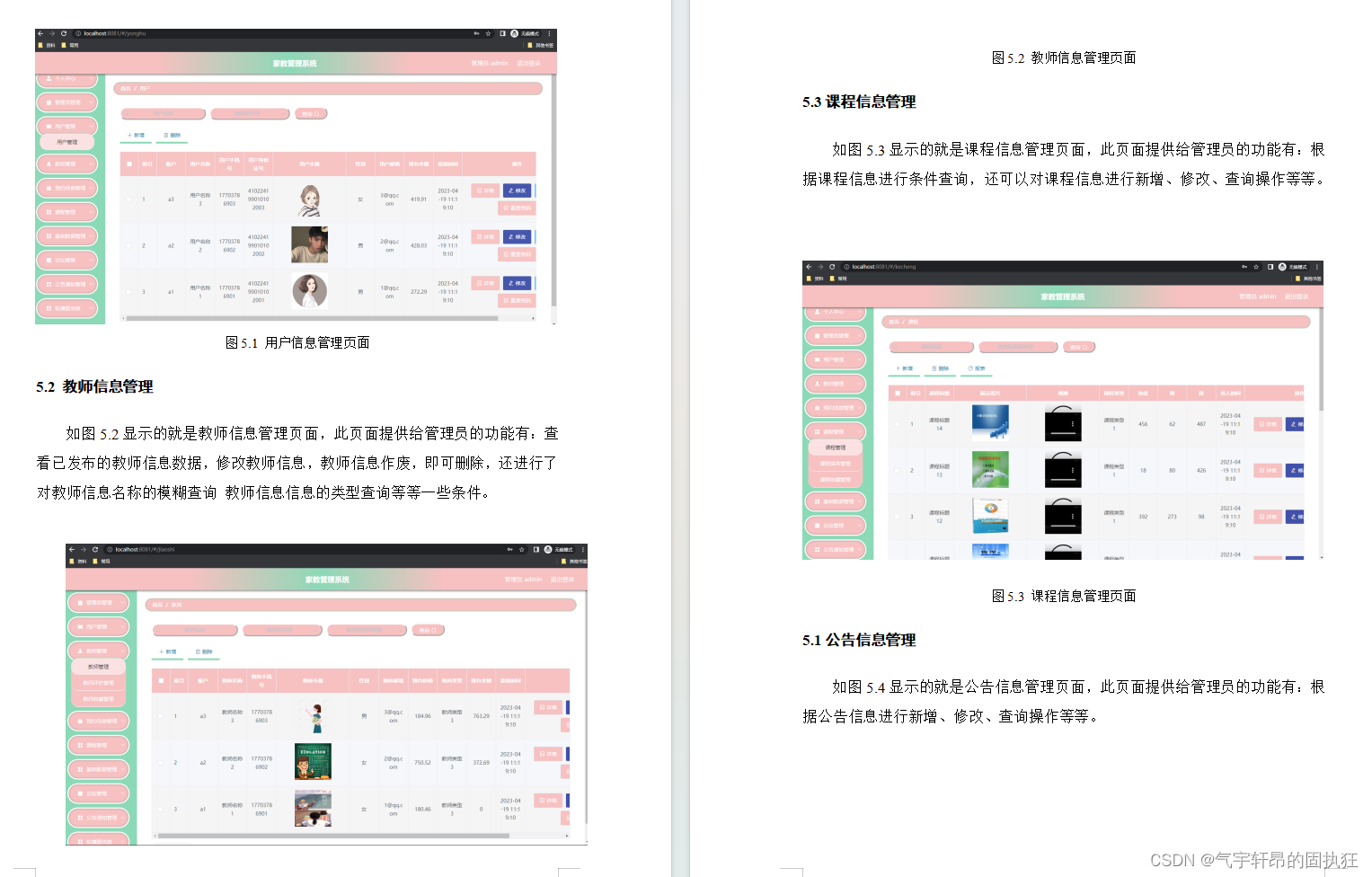
安装之前可以先了解一下论文的主要内容,便于之后网络训练与推理,调试程序。
论文地址:nnU-Net: a self-configuring method for deep learning-based biomedical image segmentation | Nature Methods
也可以从其他博客快速浏览:论文解读- nnU-Net: Self-adapting Framework for U-Net-Based Medical Image Segmentation(附实现教程)_nnunet self adaptinng-CSDN博客
如果想跟官网github一样在ubuntu下安装参考:nnUNet保姆级使用教程!从环境配置到训练与推理(新手必看)-CSDN博客
1.本博客是在win11安装,前期Anaconda的虚拟环境自己配置好,然后下载好nnUnet v1的安装包,然后解压在该目中运行:
pip install -e .
如果需要观察模型的网络结构图可以安装hiddenlayer:
nnUNet给出的指令:
pip install --upgrade git+https://github.com/FabianIsensee/hiddenlayer.git@more_plotted_details#egg=hiddenlayer上面指令自己运行报错:
× git clone --filter=blob:none --quiet https://github.com/FabianIsensee/hiddenlayer.git 'C:\Users\Administrator\AppData\Local\Temp\pip-install-dh84s7ac\hiddenlayer_be2e7545caf44fbeae10f5b0cfd81e30' did not run successfully,可能是网络原因。干脆直接从hiddenlayer官网的指令进行安装(顺利安装):
pip install git+https://github.com/waleedka/hiddenlayer.git 2.开始准备推理的数据,注意它的格式要求,开始体验一下如何用官网模型进行infer使用:
2.开始准备推理的数据,注意它的格式要求,开始体验一下如何用官网模型进行infer使用:
查看nnUNet提供的预训练好的模型:
nnUNet_print_available_pretrained_modelsTask001_BrainTumour
Brain Tumor Segmentation.
Segmentation targets are edema, enhancing tumor and necrosis,
Input modalities are 0: FLAIR, 1: T1, 2: T1 with contrast agent, 3: T2.
Also see Medical Segmentation Decathlon, http://medicaldecathlon.com/Task002_Heart
Left Atrium Segmentation.
Segmentation target is the left atrium,
Input modalities are 0: MRI.
Also see Medical Segmentation Decathlon, http://medicaldecathlon.com/Task003_Liver
Liver and Liver Tumor Segmentation.
Segmentation targets are liver and tumors,
Input modalities are 0: abdominal CT scan.
Also see Medical Segmentation Decathlon, http://medicaldecathlon.com/Task004_Hippocampus
Hippocampus Segmentation.
Segmentation targets posterior and anterior parts of the hippocampus,
Input modalities are 0: MRI.
Also see Medical Segmentation Decathlon, http://medicaldecathlon.com/Task005_Prostate
Prostate Segmentation.
Segmentation targets are peripheral and central zone,
Input modalities are 0: T2, 1: ADC.
Also see Medical Segmentation Decathlon, http://medicaldecathlon.com/Task006_Lung
Lung Nodule Segmentation.
Segmentation target are lung nodules,
Input modalities are 0: abdominal CT scan.
Also see Medical Segmentation Decathlon, http://medicaldecathlon.com/Task007_Pancreas
Pancreas Segmentation.
Segmentation targets are pancras and pancreas tumor,
Input modalities are 0: abdominal CT scan.
Also see Medical Segmentation Decathlon, http://medicaldecathlon.com/Task008_HepaticVessel
Hepatic Vessel Segmentation.
Segmentation targets are hepatic vesels and liver tumors,
Input modalities are 0: abdominal CT scan.
Also see Medical Segmentation Decathlon, http://medicaldecathlon.com/Task009_Spleen
Spleen Segmentation.
Segmentation target is the spleen,
Input modalities are 0: abdominal CT scan.
Also see Medical Segmentation Decathlon, http://medicaldecathlon.com/Task010_Colon
Colon Cancer Segmentation.
Segmentation target are colon caner primaries,
Input modalities are 0: CT scan.
Also see Medical Segmentation Decathlon, http://medicaldecathlon.com/Task017_AbdominalOrganSegmentation
Multi-Atlas Labeling Beyond the Cranial Vault - Abdomen.
Segmentation targets are thirteen different abdominal organs,
Input modalities are 0: abdominal CT scan.
Also see https://www.synapse.org/#!Synapse:syn3193805/wiki/217754Task024_Promise
Prostate MR Image Segmentation 2012.
Segmentation target is the prostate,
Input modalities are 0: T2.
Also see https://promise12.grand-challenge.org/Task027_ACDC
Automatic Cardiac Diagnosis Challenge.
Segmentation targets are right ventricle, left ventricular cavity and left myocardium,
Input modalities are 0: cine MRI.
Also see https://acdc.creatis.insa-lyon.fr/Task029_LiTS
Liver and Liver Tumor Segmentation Challenge.
Segmentation targets are liver and liver tumors,
Input modalities are 0: abdominal CT scan.
Also see https://competitions.codalab.org/competitions/17094Task035_ISBILesionSegmentation
Longitudinal multiple sclerosis lesion segmentation Challenge.
Segmentation target is MS lesions,
input modalities are 0: FLAIR, 1: MPRAGE, 2: proton density, 3: T2.
Also see https://smart-stats-tools.org/lesion-challengeTask038_CHAOS_Task_3_5_Variant2
CHAOS - Combined (CT-MR) Healthy Abdominal Organ Segmentation Challenge (Task 3 & 5).
Segmentation targets are left and right kidney, liver, spleen,
Input modalities are 0: T1 in-phase, T1 out-phase, T2 (can be any of those)
Also see https://chaos.grand-challenge.org/Task048_KiTS_clean
Kidney and Kidney Tumor Segmentation Challenge. Segmentation targets kidney and kidney tumors, Input modalities are 0: abdominal CT scan. Also see https://kits19.grand-challenge.org/Task055_SegTHOR
SegTHOR: Segmentation of THoracic Organs at Risk in CT images.
Segmentation targets are aorta, esophagus, heart and trachea,
Input modalities are 0: CT scan.
Also see https://competitions.codalab.org/competitions/21145Task061_CREMI
MICCAI Challenge on Circuit Reconstruction from Electron Microscopy Images (Synaptic Cleft segmentation task).
Segmentation target is synaptic clefts,
Input modalities are 0: serial section transmission electron microscopy of neural tissue.
Also see https://cremi.org/Task075_Fluo_C3DH_A549_ManAndSim
Fluo-C3DH-A549-SIM and Fluo-C3DH-A549 datasets of the cell tracking challenge. Segmentation target are C3DH cells in fluorescence microscopy images.
Input modalities are 0: fluorescence_microscopy
Also see http://celltrackingchallenge.net/Task076_Fluo_N3DH_SIM
Fluo-N3DH-SIM dataset of the cell tracking challenge. Segmentation target are N3DH cells and cell borders in fluorescence microscopy images.
Input modalities are 0: fluorescence_microscopy
Also see http://celltrackingchallenge.net/
Note that the segmentation output of the models are cell center and cell border. These outputs mus tbe converted to an instance segmentation for the challenge.
See https://github.com/MIC-DKFZ/nnUNet/blob/master/nnunet/dataset_conversion/Task076_Fluo_N3DH_SIM.pyTask082_BraTS2020
Brain tumor segmentation challenge 2020 (BraTS)
Segmentation targets are 0: background, 1: edema, 2: necrosis, 3: enhancing tumor
Input modalities are 0: T1, 1: T1ce, 2: T2, 3: FLAIR (MRI images)
Also see https://www.med.upenn.edu/cbica/brats2020/Task089_Fluo-N2DH-SIM_thickborder_time
Fluo-N2DH-SIM dataset of the cell tracking challenge. Segmentation target are nuclei of N2DH cells and cell borders in fluorescence microscopy images.
Input modalities are 0: t minus 4, 0: t minus 3, 0: t minus 2, 0: t minus 1, 0: frame of interest
Note that the input channels are different time steps from a time series acquisition
Note that the segmentation output of the models are cell center and cell border. These outputs mus tbe converted to an instance segmentation for the challenge.
See https://github.com/MIC-DKFZ/nnUNet/blob/master/nnunet/dataset_conversion/Task089_Fluo-N2DH-SIM.py
Also see http://celltrackingchallenge.net/Task114_heart_MNMs
Cardiac MRI short axis images from the M&Ms challenge 2020.
Input modalities are 0: MRI
See also https://www.ub.edu/mnms/
Note: Labels of the M&Ms Challenge are not in the same order as for the ACDC challenge.
See https://github.com/MIC-DKFZ/nnUNet/blob/master/nnunet/dataset_conversion/Task114_heart_mnms.pyTask115_COVIDSegChallenge
Covid lesion segmentation in CT images. Data originates from COVID-19-20 challenge.
Predicted labels are 0: background, 1: covid lesion
Input modalities are 0: CT
See also https://covid-segmentation.grand-challenge.org/Task135_KiTS2021
Kidney and kidney tumor segmentation in CT images. Data originates from KiTS2021 challenge.
Predicted labels are 0: background, 1: kidney, 2: tumor, 3: cyst
Input modalities are 0: CT
See also https://kits21.kits-challenge.org/Task169_BrainTumorPET
Brain tumor segmentation in FET PET images. Data originates from the Research Center Jülich, Germany.
Predicted labels are 0: background, 1: tumor
Input modalities are 0: FET PET
See also (NOT YET AVAILABLE)需要类似于ubuntu系统下一样,设置环境临时变量:
set RESULTS_FOLDER=自己的硬盘根目录\nnUNet-nnunetv1\dataset\nnUNet_trained_models
set nnUNet_raw_data_base=自己的硬盘根目录\nnUNet-nnunetv1\dataset\nnUNet_raw
set nnUNet_preprocessed=自己的硬盘根目录\nnUNet-nnunetv1\dataset\nnUNet_preprocessed是否设置成功,可以通过
echo %RESULTS_FOLDER%
echo %nnUNet_raw_data_base%
echo %nnUNet_preprocessed%3. 推理之前需要将数据按照nnUNet要求进行格式转换:
由于ubuntu跟win11系统路径格式不一样(不懂可以看下区别),需要提前修改相应的程序:
1)nnUNet_convert_decathlon_task.py中22行的“folder.split('/')[-1]”改成“folder.split('\\')[-1]”。
2)utils.py中40行的input_folder.split("/")[-1]改成input_folder.split("\\")[-1]
3) common_utils.py中26行的filename.split("/")[-1]改成filename.split("\\")[-1]
然后,根据数据格式转换说明:
nnUNet_convert_decathlon_task -i FOLDER_TO_TASK_AS_DOWNLOADED_FROM_MSD -p NUM_PROCESSES具体指令:
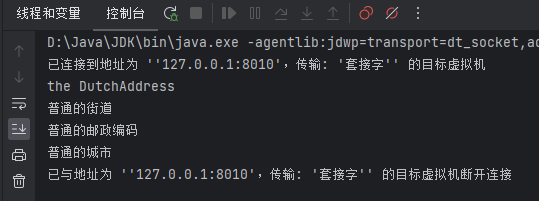
nnUNet_convert_decathlon_task -i 自己存放数据集的路径\Task05_Prostate运行结果目录如下:
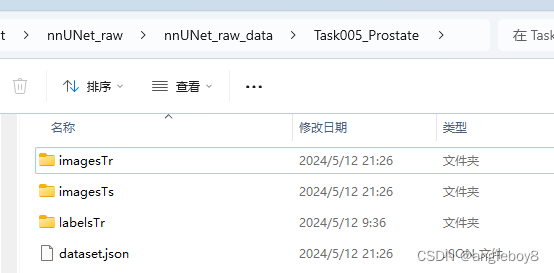
以imagesTs为例:

按照官网进行数据预处理:
nnUNet_plan_and_preprocess -t XXX --verify_dataset_integrityXXX表示任务号;
nnUNet_plan_and_preprocess -t 005 --verify_dataset_integrity4. 运行推理程序:
范例:
nnUNet_predict -i INPUT_FOLDER -o OUTPUT_FOLDER -t TASK_NAME_OR_ID
-m CONFIGURATION --save_npz这里需要注意一定要修改cropping.py中123行的i.split("/")[-1][:-4],将其修改为i.split("\\")[-1][:-4]。否则会报错误(这个错误很隐蔽,网上查了很多资料没有解决,一句句打断点找到的问题所在,debug不易,趟掉很多坑):
“return _nx.concatenate(arrs, 0, dtype=dtype, casting=casting)
ValueError: need at least one array to concatenate”检测一下nnUNet_cropped_data是否生成了预处理的文件(*.npz和*.pkl):
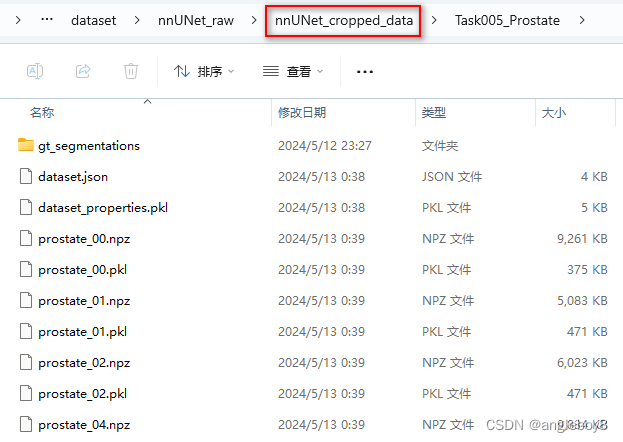
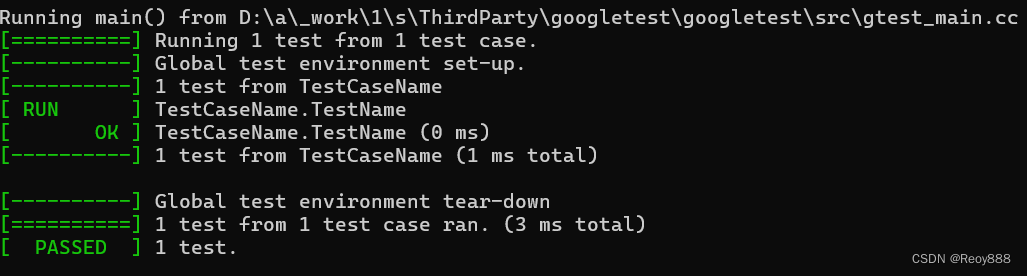
nnUNet_predict -i 自己的路径\nnUNet_raw\nnUNet_raw_data\Task005_Prostate\imagesTs -o 自己的路径\nnUNet_raw\nnUNet_raw_data\Task005_Prostate\inferTs -t 5 -m 3d_fullres -f 0运行结果:
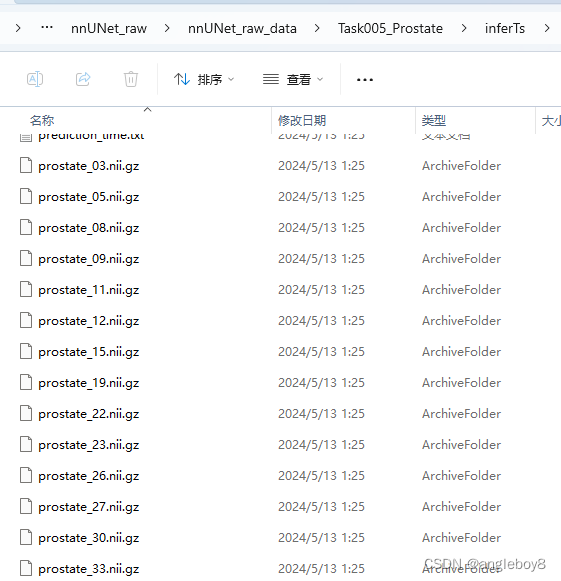
推理后得到标注结果:

现在已基本掌握了如何利用nnUNet以后模型进行推理,先会用再学如何利用自己的数据进行训练,之后重点讲如何自定义训练。这篇博客主要是为了解决nnUNet如何在win11环境中解决数据转换和数据预处理,以及如何模型推理。目前国内win11环境安装配置为此独一份,原创来之不易,点赞收藏,后期更精彩。